yield agent type
1 1.05 1 None
2 1.09 1 None
3 1.32 1 None
4 1.13 1 None
5 1.13 1 None
6 1.33 1 None
7 1.37 2 Bio1
8 1.12 2 Bio1
9 1.20 2 Bio1
10 1.24 2 Bio1
11 1.47 2 Bio1
12 1.35 2 Bio1
13 1.36 3 Bio2
14 1.33 3 Bio2
15 1.27 3 Bio2
16 1.50 3 Bio2
17 1.37 3 Bio2
18 1.14 3 Bio2
19 1.64 4 Chm1
20 1.41 4 Chm1
21 1.30 4 Chm1
22 1.46 4 Chm1
23 1.31 4 Chm1
24 1.37 4 Chm1
25 1.45 5 Chm2
26 1.34 5 Chm2
27 1.61 5 Chm2
28 1.54 5 Chm2
29 1.40 5 Chm2
30 1.66 5 Chm225 Multiple Comparison*
25.1 Multiplicity of Hypotheses
What do we do after ANOVA? In ANOVA we test
If we reject
So after ANOVA, we may want to test
-
We will be testing many hypotheses. As we discussed before, if there are
However, performing too many pairewise tests simultaneously is problematic. If there are 10 hypotheses and each is tested at
This high probability problem always arises due to multiplicity of hypotheses, meaning that multiple testings are conducted simultaneously in one single research study. This problem of multiplicity is more serious when you are testing more hypotheses.
[Example: Gene Expression Analysis] A data set is collected on gene expressions on 10,000 genes. This kind of research is done if you are trying to find genes responsible for cancer. If those genes are correctly detected, you can study their structure and come up with a cure.
If we test each of the hypotheses at
In this chapter we will learn how to do multiple pairwise comparisons or testings simultaneously while controlling the familywise error rate at a low level, reducing the number of false positives, or mitigating the inflated Type-I error rate due to multiple comparisons. The main methods introduced here include Bonferroni correction, Fisher’s LSD (Least Significant Difference), Tukey’s HSD (honestly significant difference), and Dunnett’s Method. Before we jump into these methods, let’s first look at an examples, refreshing our knowledge of ANOVA.
Example of Multiple Comparison: ANOVA (Example 9.4 in SMD)
A study was done to test 5 different agents used to control weeds. Each of these agents were applied to sample of 6 one-acre plots. The hay was harvested and the total yield was recorded.
The data set is saved in the R data frame data_weed.
Summary statistics are shown below.
agent sample_mean sample_sd sample_size type
1 1.17 0.120 6 None
2 1.29 0.127 6 Bio1
3 1.33 0.120 6 Bio2
4 1.42 0.125 6 Chm1
5 1.50 0.127 6 Chm2The data set is saved in the Python data frame data_weed.
import pandas as pd
# Load the data from a CSV file
data_weed = pd.read_csv("./data/data_weed.csv")
data_weed yield agent type
0 1.0480 1 NaN
1 1.0896 1 NaN
2 1.3151 1 NaN
3 1.1275 1 NaN
4 1.1349 1 NaN
5 1.3348 1 NaN
6 1.3659 2 Bio1
7 1.1238 2 Bio1
8 1.2049 2 Bio1
9 1.2387 2 Bio1
10 1.4730 2 Bio1
11 1.3517 2 Bio1
12 1.3621 3 Bio2
13 1.3342 3 Bio2
14 1.2703 3 Bio2
15 1.4950 3 Bio2
16 1.3714 3 Bio2
17 1.1350 3 Bio2
18 1.6369 4 Chm1
19 1.4142 4 Chm1
20 1.3014 4 Chm1
21 1.4625 4 Chm1
22 1.3093 4 Chm1
23 1.3657 4 Chm1
24 1.4532 5 Chm2
25 1.3362 5 Chm2
26 1.6145 5 Chm2
27 1.5390 5 Chm2
28 1.3967 5 Chm2
29 1.6603 5 Chm2 agent sample_mean sample_sd sample_size type
1 1.17 0.120 6 None
2 1.29 0.127 6 Bio1
3 1.33 0.120 6 Bio2
4 1.42 0.125 6 Chm1
5 1.50 0.127 6 Chm2The questions are
- if there is a difference between the agents
- which agent provides the best yield
The first question can be answered by ANOVA. Let’s check the assumptions, and perform ANOVA.
First we check the equality of variances
library(car)
(levene_test <- leveneTest(yield ~ agent, data = data_weed))Levene's Test for Homogeneity of Variance (center = median)
Df F value Pr(>F)
group 4 0.12 0.97
25 # Bartlett test is adapted for normally distributed data.
(bartlett_test <- bartlett.test(yield ~ agent, data = data_weed))
Bartlett test of homogeneity of variances
data: yield by agent
Bartlett's K-squared = 0.03, df = 4, p-value = 1# The Fligner-Killeen test is most robust against departures from normality. Use it when there are outliers.
(fligner_test <- fligner.test(yield ~ agent, data = data_weed))
Fligner-Killeen test of homogeneity of variances
data: yield by agent
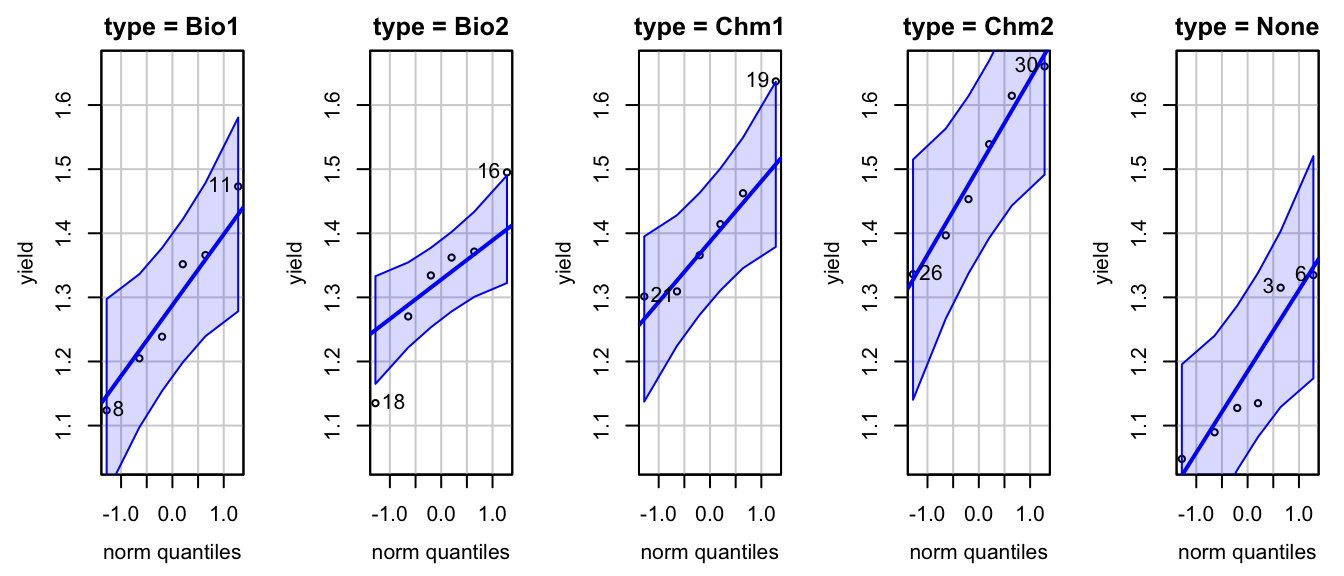
Fligner-Killeen:med chi-squared = 0.4, df = 4, p-value = 1Then we check the normality assumption
import scipy.stats as stats
import statsmodels.api as sm
from statsmodels.formula.api import ols
# Levene's Test
# https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.levene.html
stats.levene(data_weed['yield'][data_weed['agent'] == 1],
data_weed['yield'][data_weed['agent'] == 2],
data_weed['yield'][data_weed['agent'] == 3],
data_weed['yield'][data_weed['agent'] == 4],
data_weed['yield'][data_weed['agent'] == 5])LeveneResult(statistic=0.11984660886763815, pvalue=0.9741358089178117)# Bartlett's Test
# https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.bartlett.html
stats.bartlett(data_weed['yield'][data_weed['agent'] == 1],
data_weed['yield'][data_weed['agent'] == 2],
data_weed['yield'][data_weed['agent'] == 3],
data_weed['yield'][data_weed['agent'] == 4],
data_weed['yield'][data_weed['agent'] == 5])BartlettResult(statistic=0.02859647293854971, pvalue=0.99989874938744)# Fligner-Killeen Test
# https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.fligner.html
stats.fligner(data_weed['yield'][data_weed['agent'] == 1],
data_weed['yield'][data_weed['agent'] == 2],
data_weed['yield'][data_weed['agent'] == 3],
data_weed['yield'][data_weed['agent'] == 4],
data_weed['yield'][data_weed['agent'] == 5])FlignerResult(statistic=0.4334833327653211, pvalue=0.9796448746915967)Then we check the normality assumption
Next, we perform ANOVA to test if there is a difference among the agents.
## need to rename yield. need to figure out why
data_weed.rename(columns={'yield': 'yield_value'}, inplace=True)
sm.stats.anova_lm(ols('yield_value ~ C(agent)', data=data_weed).fit()) df sum_sq mean_sq F PR(>F)
C(agent) 4.0 0.364675 0.091169 5.958351 0.001647
Residual 25.0 0.382525 0.015301 NaN NaNWe reject
In the ANOVA, we only conclude that mean yields are different under different agents. How do we say which agent is the best? Are chemical agents (Agent 4, 5) better than the biological agents (Agent 2, 3)? All of these questions can be answered by all pairwise comparisons.
Here shows the boxplot for each weed agent. The ANOVA has told us there are some mean yield differences among these agents, and we are going to find out which 2 agents or which pair produce statistically discernible different mean yields.
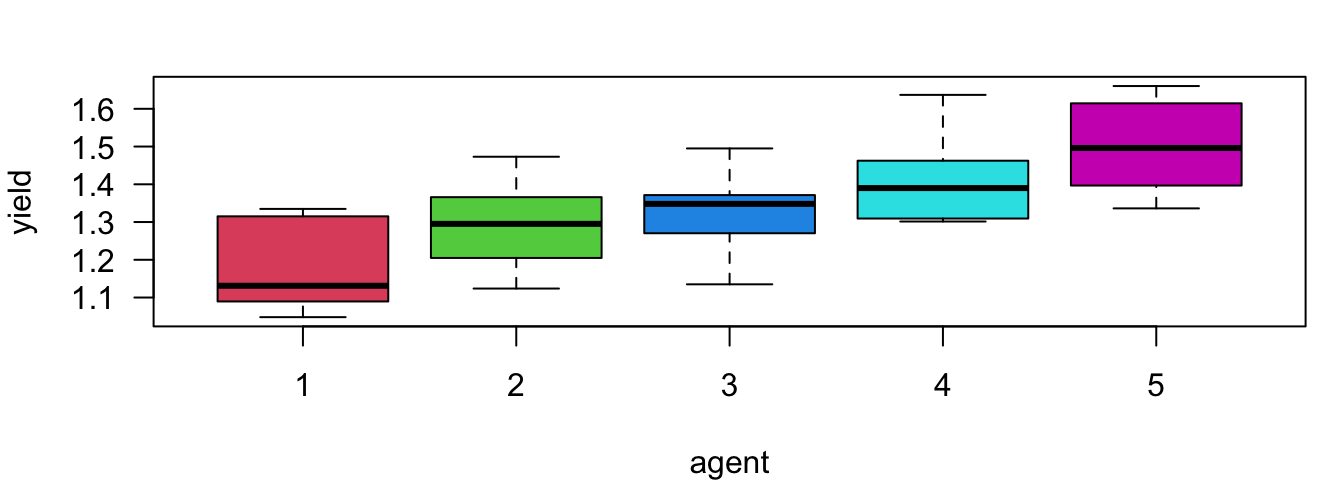
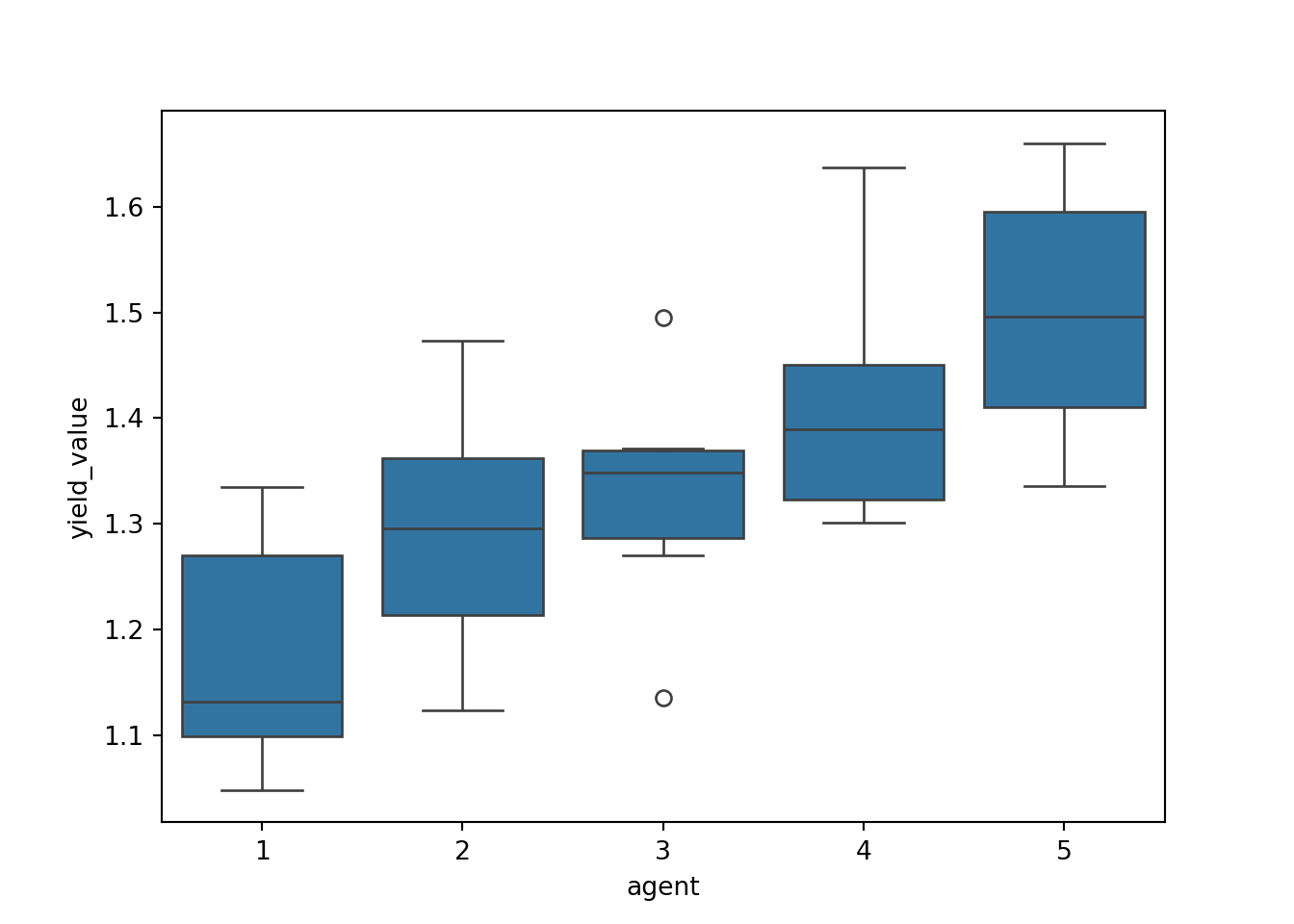
25.2 Familywise Error Rate (FWER) and Bonferroni Correction
25.2.1 Familywise Error Rate (FWER)
Instead of using Type I error rate
The idea is that when we are doing multiple comparisons, we view all the comparisons, here in the weed yield example, all 10 2-sample tests, as a whole single problem like one single family, and we are controlling the falsely rejection rate for that one family, not the falsely rejection rate for every single family member, a 2 sample test in the family.
If there are
As shown in the figure below, if
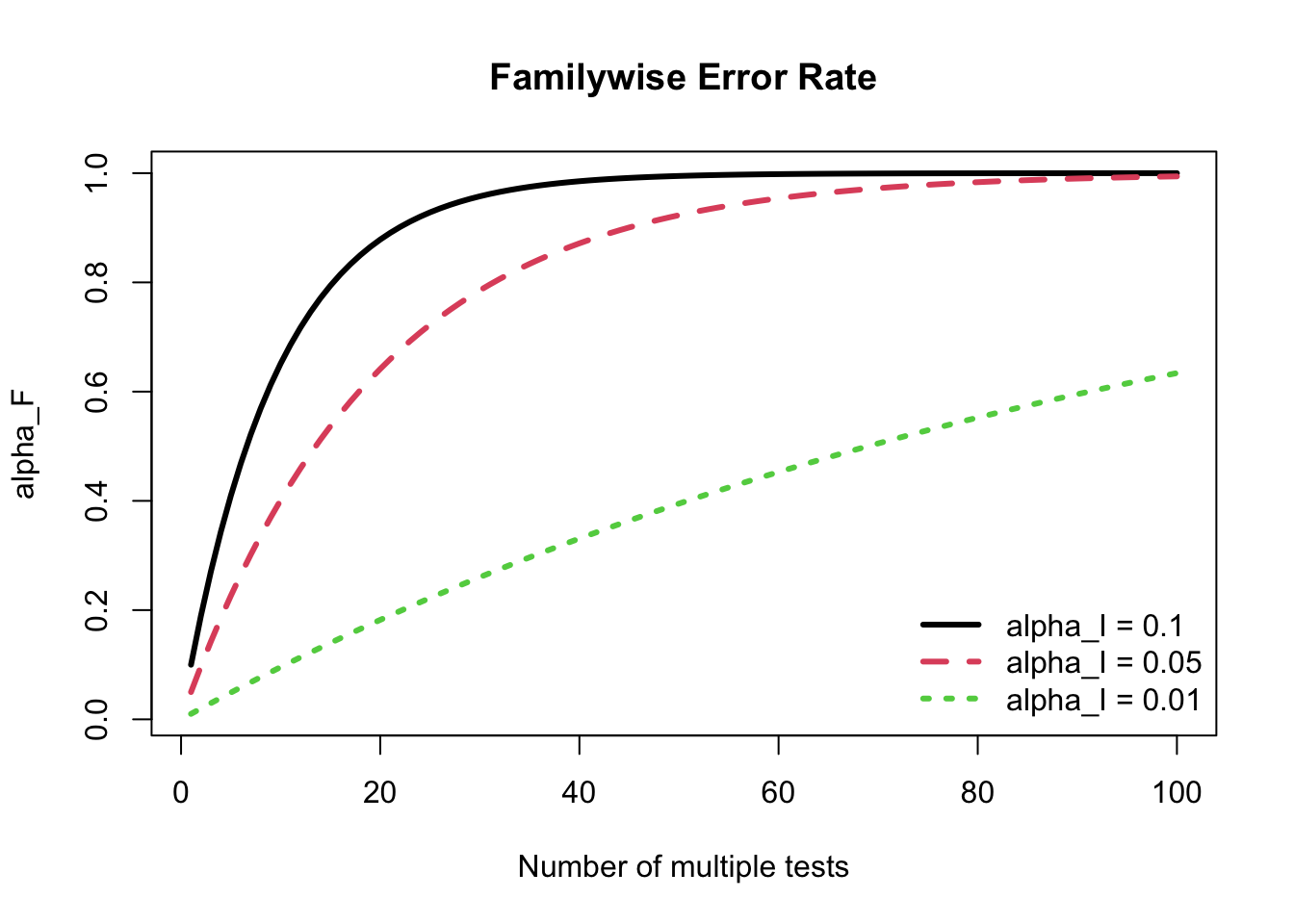
25.2.2 Bonferroni Correction
We will discuss four methods: Bonferroni, Fisher’s LSD (Least Significant Difference), Tukey’s HSD (Honestly Significant Difference), and Dunnett’s method. With
The idea is that when
The problem is the threshold
The first method is Bonferroni correction. The Bonferroni inequality provides a method for selecting
If there are
However, the problem with this approach is that if
In the Bonferroni method, we use test statistic
The value
Look at the formula carefully. The differences between Bonferroni and the unadjusted 2-sample pooled
- Change the critical value from
- Change the pooled variance from
In Bonferroni method, we make the individual error rate smaller, and we consider all groups when estimating
Example of Multiple Comparison: Bonferroni
agent sample_mean sample_sd sample_size type
1 1.17 0.120 6 None
2 1.29 0.127 6 Bio1
3 1.33 0.120 6 Bio2
4 1.42 0.125 6 Chm1
5 1.50 0.127 6 Chm2Back to the weed yield example, and see the pair comparison result using the Bonferroni method. We have 5 different weed agents. One is control group, no agent (Agent 1), 2 biological agents (Agent 2 and 3), and 2 chemical agents (Agent 4 and 5).
We know
Also, set
-
Therefore, using the Bonferroni method, we conclude
Here lists comparisons between Agent 1 (control) with Agent 2 to 5.
- Agent 1 v.s. 5:
- Agent 1 v.s. 4:
- Agent 1 v.s. 3:
- Agent 1 v.s. 2:
We can see that Chm2 (Agent 5) and Chm1 (Agent 4) are different from Control (Agent 1), but Bio1 and Bio2 are not significantly different from the control group.
Let me first show how I do the Bonferroni correction using R step by step.
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 1.18 1.18 1.18 1.18 1.29 1.29 1.29 1.33 1.33 1.42
[2,] 1.29 1.33 1.42 1.50 1.33 1.42 1.50 1.42 1.50 1.50## all pair distances |y_bar_i - y_bar_j|
y_bar_dist <- apply(y_bar_pair, 2, dist)
dd <- t(combn(1:5, 2))
names(y_bar_dist) <- paste(dd[, 1], dd[, 2], sep = " vs ")
y_bar_dist1 vs 2 1 vs 3 1 vs 4 1 vs 5 2 vs 3 2 vs 4 2 vs 5 3 vs 4 3 vs 5 4 vs 5
0.118 0.153 0.240 0.325 0.035 0.122 0.207 0.087 0.172 0.085 ## set values
alpha <- 0.05; m <- choose(5, 2); n <- 6
## perform anova
aov_res <- aov(yield ~ agent, data = data_weed)
## extract df_W
(df <- df.residual(aov_res)) [1] 25## extract s_W
(sw <- sigma(aov_res) )[1] 0.124## critical value of bonferroni
(t_bon <- qt(p = alpha / (2 * m), df = df, lower.tail = FALSE))[1] 3.08## margin of error
(E_bon <- t_bon * sw * sqrt((1/n + 1/n)))[1] 0.22## decision
(y_bar_dist > E_bon) 1 vs 2 1 vs 3 1 vs 4 1 vs 5 2 vs 3 2 vs 4 2 vs 5 3 vs 4 3 vs 5 4 vs 5
FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE There is a shortcut to do Bonferroni correction for pairwise comparisons. We can use the function pairwise.t.test(). If we want to do 2-sample pooled pool.sd should be FALSE because this argument is for the pooled variance pooled by all groups combined, not group p.adjust.method is none because we do not control or adjust FWER. The var.equal should be TRUE, so that the pooled
The output is a matrix including p-values of all pair comparison testings. We can tell which pairs are significant by comparing their p-value with the significance level
If we want to do Bonferroni correction, use p.adjust.method = "bonferroni". Again, in Bonferroni, (1, 4) and (1, 5) are statistically significantly different. In traditional 2 sample
## individual 2-sample pooled t-test but not pooled by all groups
pairwise.t.test(data_weed$yield, data_weed$agent, pool.sd = FALSE,
p.adjust.method = "none", var.equal = TRUE)
Pairwise comparisons using t tests with non-pooled SD
data: data_weed$yield and data_weed$agent
1 2 3 4
2 0.129 - - -
3 0.052 0.634 - -
4 0.007 0.124 0.246 -
5 0.001 0.018 0.036 0.269
P value adjustment method: none ## Bonferroni correction
pairwise.t.test(data_weed$yield, data_weed$agent,
p.adjust.method = "bonferroni")
Pairwise comparisons using t tests with pooled SD
data: data_weed$yield and data_weed$agent
1 2 3 4
2 1.000 - - -
3 0.421 1.000 - -
4 0.025 1.000 1.000 -
5 0.001 0.077 0.237 1.000
P value adjustment method: bonferroni Let me first show how I do the Bonferroni correction using Python step by step.
import itertools
# Given data
y_bar = np.array([1.175, 1.293, 1.328, 1.415, 1.500])
# All pairs
y_bar_pair = np.array(list(itertools.combinations(y_bar, 2)))
y_bar_pairarray([[1.175, 1.293],
[1.175, 1.328],
[1.175, 1.415],
[1.175, 1.5 ],
[1.293, 1.328],
[1.293, 1.415],
[1.293, 1.5 ],
[1.328, 1.415],
[1.328, 1.5 ],
[1.415, 1.5 ]])# All pair distances |y_bar_i - y_bar_j|
y_bar_dist = np.abs(y_bar_pair[:, 0] - y_bar_pair[:, 1])
y_bar_distarray([0.118, 0.153, 0.24 , 0.325, 0.035, 0.122, 0.207, 0.087, 0.172,
0.085])# Combination indices (for naming)
dd = np.array(list(itertools.combinations(range(1, 6), 2)))
# Create names for the distances
y_bar_dist_names = [f"{i} vs {j}" for i, j in dd]
# Display the distances with names
y_bar_dist_dict = dict(zip(y_bar_dist_names, y_bar_dist))
y_bar_dist_dict{'1 vs 2': 0.11799999999999988, '1 vs 3': 0.15300000000000002, '1 vs 4': 0.24, '1 vs 5': 0.32499999999999996, '2 vs 3': 0.03500000000000014, '2 vs 4': 0.12200000000000011, '2 vs 5': 0.20700000000000007, '3 vs 4': 0.08699999999999997, '3 vs 5': 0.17199999999999993, '4 vs 5': 0.08499999999999996}from scipy.stats import t
import math
import statsmodels.api as sm
from statsmodels.formula.api import ols
# Set values
alpha = 0.05
m = math.comb(5, 2) # Equivalent to choose(5, 2)
n = 6
# Assuming 'data_weed' is your DataFrame and 'yield' and 'agent' are column names
# Perform ANOVA
data_weed['agent'] = data_weed['agent'].astype(str)
model_fit = ols('yield_value ~ agent', data=data_weed).fit()
anova_table = sm.stats.anova_lm(model_fit)
# Extract df_W (degrees of freedom) and s_W (residual standard deviation)
df = model_fit.df_resid
df25.0sw = np.sqrt(anova_table['mean_sq'].iloc[1])
sw0.12369717161951066# Critical value for Bonferroni correction
t_bon = t.ppf(1 - alpha / (2 * m), df)
t_bon3.078199460536119# Margin of error
E_bon = t_bon * sw * np.sqrt((1/n + 1/n))
E_bon0.2198345252258888# Assuming y_bar_dist was already calculated (from previous code)
# Decision
y_bar_dist > E_bonarray([False, False, True, True, False, False, False, False, False,
False])If we don’t do any correction, we can do pairwise t test for all pairs directly using ttest_ind() that conducts t-test for the means of two independent samples. It is brutal force, and there should be a better way to conduct all pairwise two-sample pooled t test separately by default.
from scipy import stats
test12 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '1', 'yield_value'],
data_weed.loc[data_weed['agent'] == '2', 'yield_value'])
test12.pvalue0.12937022290977485test13 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '1', 'yield_value'],
data_weed.loc[data_weed['agent'] == '3', 'yield_value'])
test13.pvalue0.051684324302770854test14 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '1', 'yield_value'],
data_weed.loc[data_weed['agent'] == '4', 'yield_value'])
test14.pvalue 0.0068949544337039625test15 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '1', 'yield_value'],
data_weed.loc[data_weed['agent'] == '5', 'yield_value'])
test15.pvalue 0.0010440332236669495test23 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '2', 'yield_value'],
data_weed.loc[data_weed['agent'] == '3', 'yield_value'])
test23.pvalue0.6335961134332914test24 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '2', 'yield_value'],
data_weed.loc[data_weed['agent'] == '4', 'yield_value'])
test24.pvalue0.12419123394098548test25 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '2', 'yield_value'],
data_weed.loc[data_weed['agent'] == '5', 'yield_value'])
test25.pvalue0.01786227373198035test34 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '3', 'yield_value'],
data_weed.loc[data_weed['agent'] == '4', 'yield_value'])
test34.pvalue0.24606446614093755test35 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '3', 'yield_value'],
data_weed.loc[data_weed['agent'] == '5', 'yield_value'])
test35.pvalue0.0360878773096397test45 = stats.ttest_ind(data_weed.loc[data_weed['agent'] == '4', 'yield_value'],
data_weed.loc[data_weed['agent'] == '5', 'yield_value'])
test45.pvalue0.2687698246654925There is a shortcut to do Bonferroni correction for pairwise comparisons. We can use the method t_test_pairwise(). The output saved in result_frame is a matrix including p-values of all pair comparison testings. We can tell which pairs are significant by comparing their p-value with the significance level
If we want to do Bonferroni correction, use method='bonferroni'. Again, in Bonferroni, (1, 4) and (1, 5) are statistically significantly different. In traditional 2 sample
pairwise_t_results = model_fit.t_test_pairwise(term_name='agent', method='bonferroni', alpha=0.05)
pairwise_t_results.result_frame coef std err ... pvalue-bonferroni reject-bonferroni
2-1 0.118017 0.071417 ... 1.000000 False
3-1 0.153017 0.071417 ... 0.420707 False
4-1 0.240017 0.071417 ... 0.024990 True
5-1 0.325000 0.071417 ... 0.001194 True
3-2 0.035000 0.071417 ... 1.000000 False
4-2 0.122000 0.071417 ... 0.999726 False
5-2 0.206983 0.071417 ... 0.076997 False
4-3 0.087000 0.071417 ... 1.000000 False
5-3 0.171983 0.071417 ... 0.237341 False
5-4 0.084983 0.071417 ... 1.000000 False
[10 rows x 8 columns]For each pair, we can draw its confidence interval for
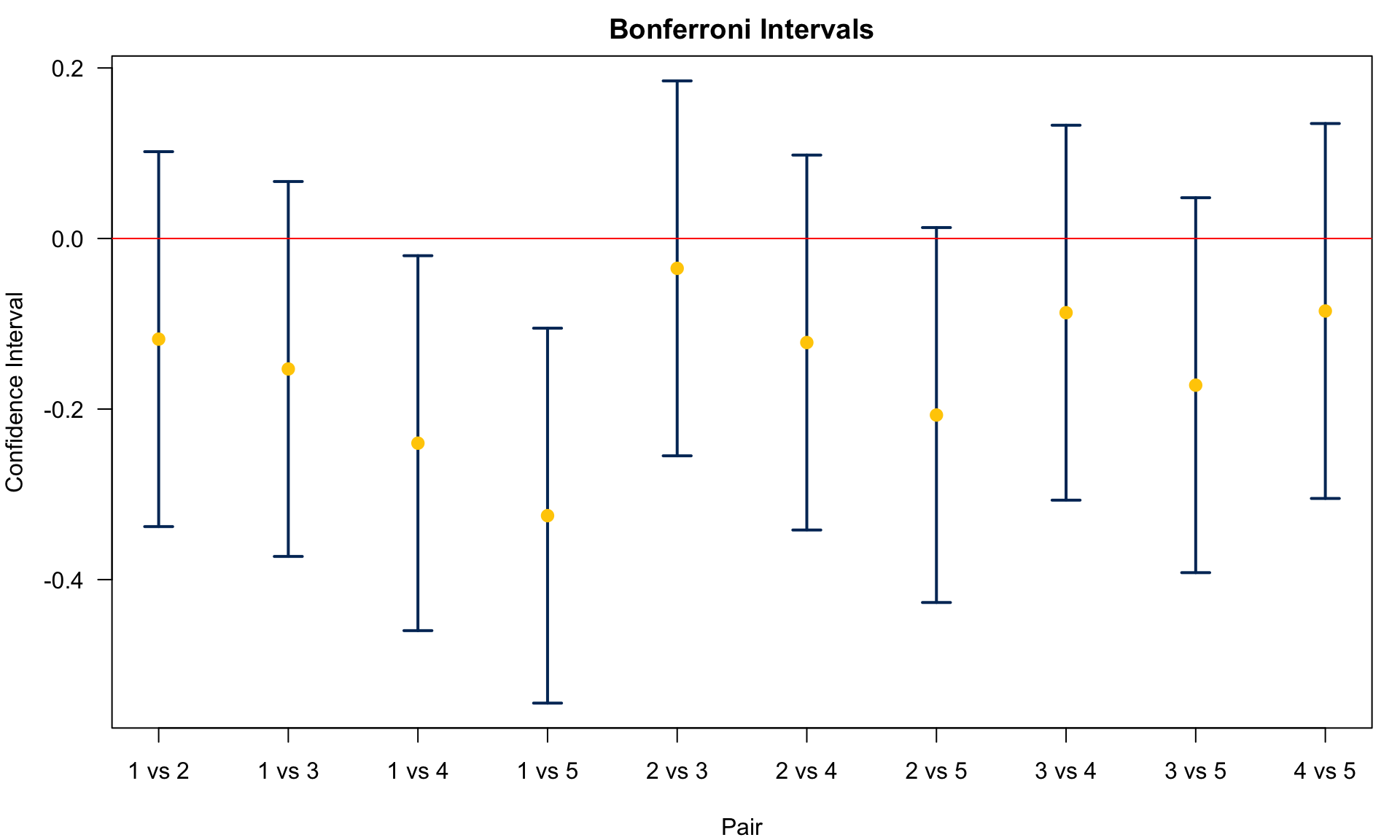
25.3 Fisher’s LSD (Least Significant Difference)
The second method introduced here is Fisher’s LSD, short for Least Significant Difference. We reject
Example of Multiple Comparison: Fisher’s LSD
agent sample_mean sample_sd sample_size type
1 1.17 0.120 6 None
2 1.29 0.127 6 Bio1
3 1.33 0.120 6 Bio2
4 1.42 0.125 6 Chm1
5 1.50 0.127 6 Chm2The process of doing Fisher’s LSD is pretty similar to doing Bonferroni.
We conclude
Agent 1 v.s. 5:
Agent 1 v.s. 4:
Agent 1 v.s. 3:
Agent 1 v.s. 2:
Agent 2 v.s. 5:
Agent 2 v.s. 4:
Agent 2 v.s. 3:
Agent 3, 4 and 5 are different from control. Agent 2 and 5 are different.
df[1] 25sw ^ 2[1] 0.0153(t_lsd <- qt(p = alpha/2, df = df, lower.tail = FALSE))[1] 2.06(E_lsd <- t_lsd * sw * sqrt(1/n + 1/n))[1] 0.147(y_bar_dist > E_lsd)1 vs 2 1 vs 3 1 vs 4 1 vs 5 2 vs 3 2 vs 4 2 vs 5 3 vs 4 3 vs 5 4 vs 5
FALSE TRUE TRUE TRUE FALSE FALSE TRUE FALSE TRUE FALSE If we use pairwise.t.test(), pool.sd = TRUE because we do use the pooled variance p.adjust.method = "none" because Fisher’s LSD does not adjust the Type I error rate. It still uses
pairwise.t.test(data_weed$yield, data_weed$agent, pool.sd = TRUE,
p.adjust.method = "none") ## Fisher’s LSD
Pairwise comparisons using t tests with pooled SD
data: data_weed$yield and data_weed$agent
1 2 3 4
2 0.111 - - -
3 0.042 0.628 - -
4 0.002 0.100 0.235 -
5 1e-04 0.008 0.024 0.245
P value adjustment method: none df25.0sw ** 20.015300990266666674t_lsd = t.ppf(1 - alpha/2, df)
t_lsd2.059538552753294E_lsd = t_lsd * sw * np.sqrt(1/n + 1/n)
E_lsd0.14708523139370552y_bar_dist > E_lsdarray([False, True, True, True, False, False, True, False, True,
False])The following should be replaced with LSD method which is still not found in any Python package. Need to update this.
pairwise_t_results = model_fit.t_test_pairwise(term_name='agent', method='bonferroni', alpha=0.05)
pairwise_t_results.result_frameThe Fisher’s LSD confidence interval for
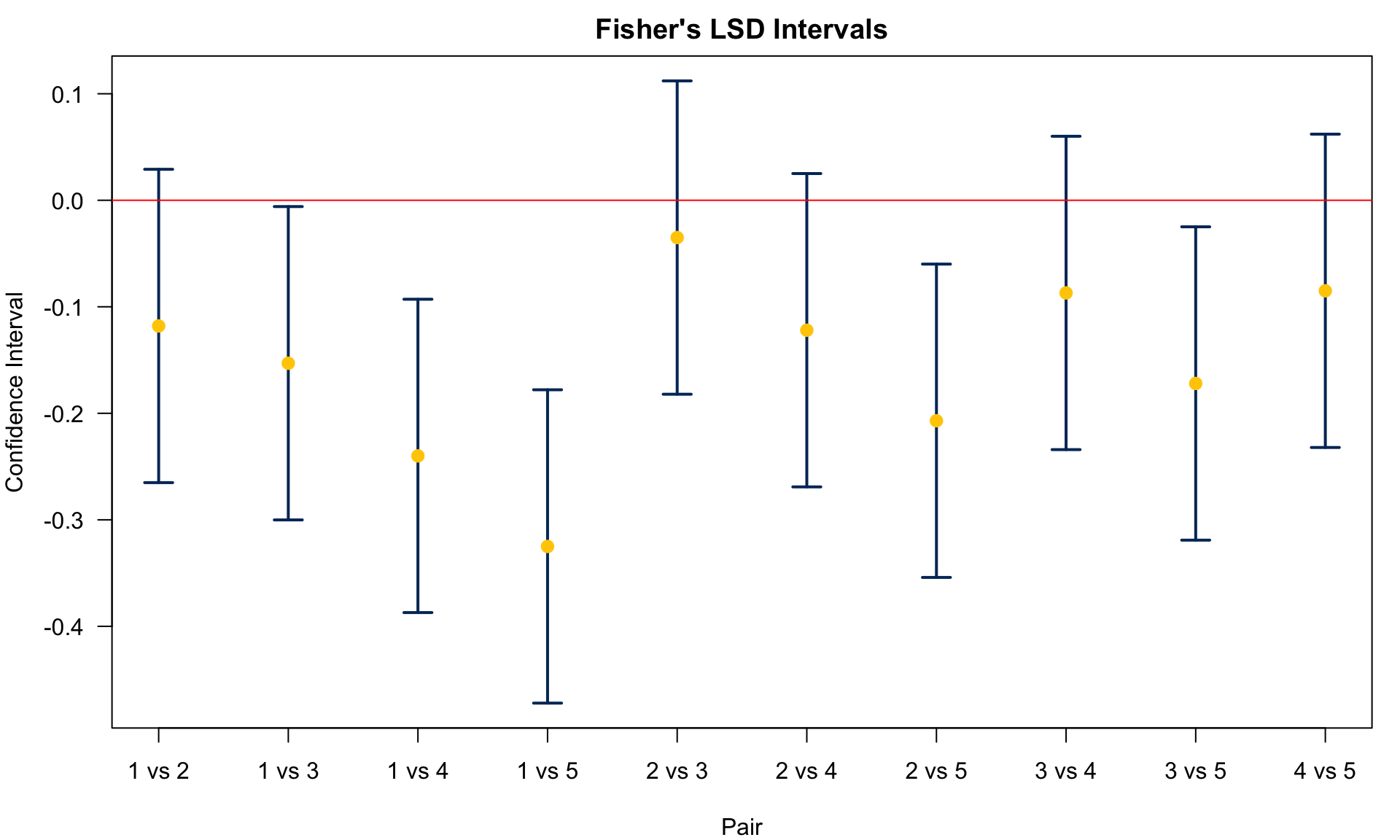
25.4 Tukey’s HSD (honestly significant difference)
The next method is the Tukey’s HSD method. We reject
Here, the square root term or the standard error keeps the same as before, but we don’t use a
Studentized range distribution is similar to
In R, we can use qtukey(p = alpha, nmeans = k, df = v, lower.tail = FALSE) to obtain
Example of Multiple Comparison: Tukey’s HSD
agent sample_mean sample_sd sample_size type
1 1.17 0.120 6 None
2 1.29 0.127 6 Bio1
3 1.33 0.120 6 Bio2
4 1.42 0.125 6 Chm1
5 1.50 0.127 6 Chm2We conclude
Agent 1 v.s. 5:
Agent 1 v.s. 4:
Agent 1 v.s. 3:
Agent 1 v.s. 2:
Agent 2 v.s. 5:
Agent 2 v.s. 4:
Agent 2 v.s. 3:
Agents 4 and 5 are different from control. No Significant difference between agents 3, 4, 5 and 2.
df[1] 25sw^2[1] 0.0153(q_tukey <- qtukey(p = alpha, nmeans = 5, df = df, lower.tail = FALSE))[1] 4.15(E_tukey <- q_tukey * sw * sqrt(1 / n))[1] 0.21(y_bar_dist > E_tukey)1 vs 2 1 vs 3 1 vs 4 1 vs 5 2 vs 3 2 vs 4 2 vs 5 3 vs 4 3 vs 5 4 vs 5
FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE A faster way to conduct Tukey’s HSD test is to use R function TukeyHSD(). After doing ANOVA, we can save the result, say res_aov, then to implement Tukey method, just put the ANOVA result in the function TukeyHSD(). It will provide all information for each pair comparison.
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = yield ~ agent, data = data_weed)
$agent
diff lwr upr p adj
2-1 0.118 -0.09172 0.328 0.480
3-1 0.153 -0.05672 0.363 0.234
4-1 0.240 0.03028 0.450 0.019
5-1 0.325 0.11526 0.535 0.001
3-2 0.035 -0.17474 0.245 0.988
4-2 0.122 -0.08774 0.332 0.447
5-2 0.207 -0.00276 0.417 0.054
4-3 0.087 -0.12274 0.297 0.741
5-3 0.172 -0.03776 0.382 0.146
5-4 0.085 -0.12476 0.295 0.757In the output, diff is lwr and upr are the lower bound and upper bound of the CI for p-adj column. With
The studentized range distribution in Python is implemented by the function studentized_range, again from scipy.stats.
df25.0sw**20.015300990266666674from scipy.stats import studentized_range
q_tukey = studentized_range.ppf(q=1-alpha, k=5, df=df)
q_tukey4.1533633299635335E_tukey = q_tukey * sw * np.sqrt(1 / n)
E_tukey0.2097413545569429y_bar_dist > E_tukeyarray([False, False, True, True, False, False, False, False, False,
False])A faster way to conduct Tukey’s HSD test is to use Python function pairwise_tukeyhsd() from statsmodels.stats.multicomp. It will provide all information for each pair comparison.
from statsmodels.stats.multicomp import pairwise_tukeyhsd
tukey = pairwise_tukeyhsd(endog=data_weed['yield_value'],
groups=data_weed['agent'], alpha=0.05)
print(tukey)Multiple Comparison of Means - Tukey HSD, FWER=0.05
===================================================
group1 group2 meandiff p-adj lower upper reject
---------------------------------------------------
1 2 0.118 0.4797 -0.0917 0.3278 False
1 3 0.153 0.2342 -0.0567 0.3628 False
1 4 0.24 0.0192 0.0303 0.4498 True
1 5 0.325 0.001 0.1153 0.5347 True
2 3 0.035 0.9876 -0.1747 0.2447 False
2 4 0.122 0.4472 -0.0877 0.3317 False
2 5 0.207 0.0543 -0.0028 0.4167 False
3 4 0.087 0.7411 -0.1227 0.2967 False
3 5 0.172 0.1461 -0.0378 0.3817 False
4 5 0.085 0.7569 -0.1248 0.2947 False
---------------------------------------------------In the output, meandiff is lower and upper are the lower bound and upper bound of the CI for
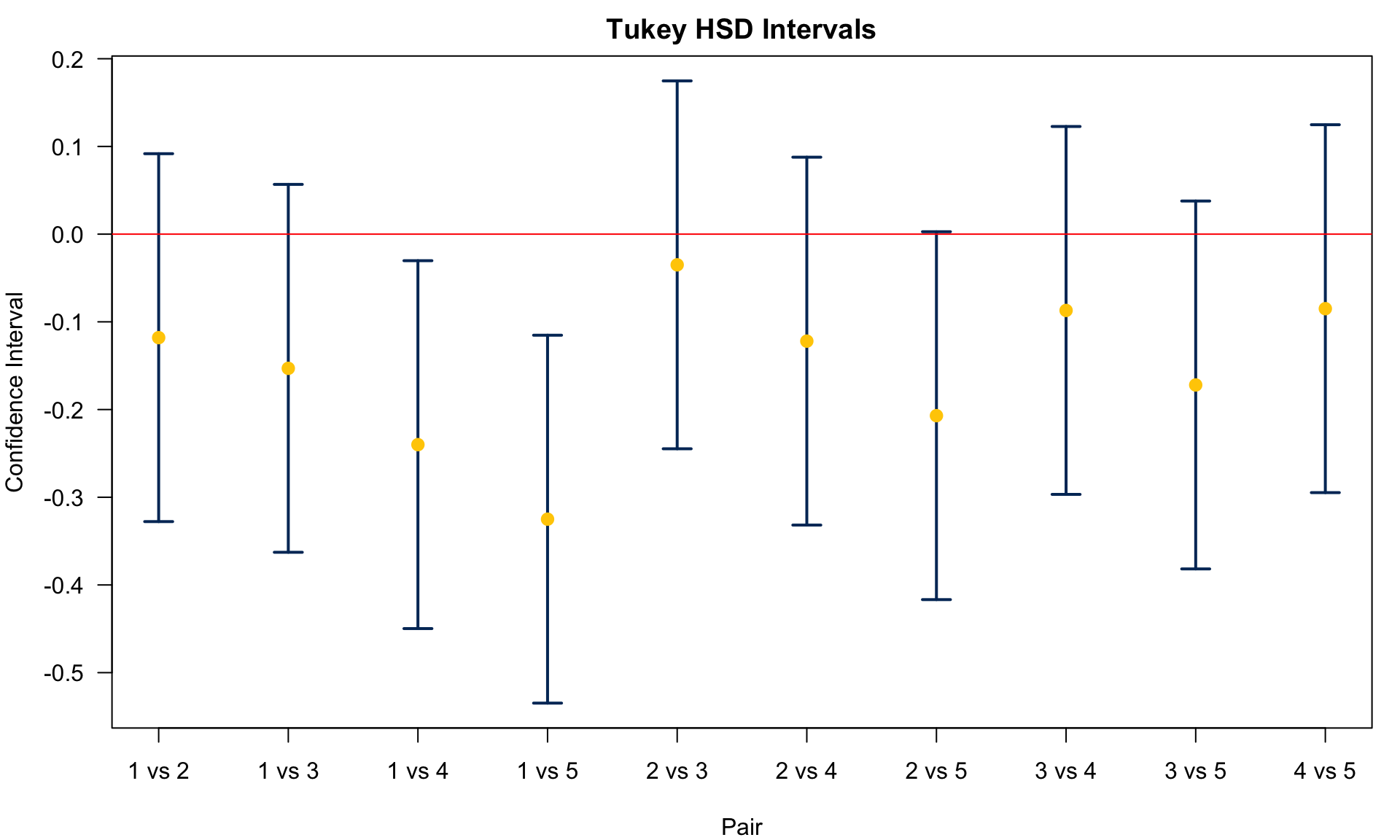
25.5 Dunnett’s Method
The last but not least method introduced here is Dunnett’s method. We reject
Well we still have the same standard error, the square root term, but we use another critical value
Note that this method is only used for comparing with a control or reference group. In our example, Agent 1 is the control group. So the parameter
Consider 2 treatment groups and one control group. If you only want to compare the 2 treatment groups with the control group, and do not want to compare the 2 treatment groups to each other, the Dunnett’s test is preferred.
Example of Multiple Comparison: Dunnett’s Method
We conclude
Agent 1 v.s. 5:
Agent 1 v.s. 4:
Agent 1 v.s. 3:
Agent 1 v.s. 2:
Only the chemical agents 4 and 5 are different from control. No biological agents are different from control.
There is no R built-in function that computes nCDunnett, and use the function qNCDun(). NC stands for Non-Central. The argument p is the probability to the left tail and keep in mind that the subscript nu is the degrees of freedom. rho here we use 0.5, meaning that all sample sizes are equal. The sample size in each group is 6, and we need to specify it 4 times corresponding to the number of non-control groups. delta is the non-centrality parameter that needs to be specified 4 times too. Here we assume they are all zero, i.e., the test uses the central distribution.
library(nCDunnett)
(d_dun <- qNCDun(p = 1 - alpha, nu = df,
rho = c(0.5, 0.5, 0.5, 0.5),
delta = c(0, 0, 0, 0), two.sided = TRUE))[1] 2.61(E_dun <- d_dun * sw * sqrt(2 / n))[1] 0.186head(y_bar_dist > E_dun, 4)1 vs 2 1 vs 3 1 vs 4 1 vs 5
FALSE FALSE TRUE TRUE DescTools.DunnettTest() performs Dunnett’s test that does multiple comparisons of means against a control group.
Attaching package: 'DescTools'The following object is masked from 'package:car':
RecodeThe following object is masked from 'package:openintro':
cardsDunnettTest(x=data_weed$yield, g=data_weed$agent)
Dunnett's test for comparing several treatments with a control :
95% family-wise confidence level
$`1`
diff lwr.ci upr.ci pval
2-1 0.118 -0.0682 0.304 0.30737
3-1 0.153 -0.0332 0.339 0.12931
4-1 0.240 0.0538 0.426 0.00845 **
5-1 0.325 0.1387 0.511 0.00043 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1There is no functions in Python that calculates probabilities and quantiles of the Dunnett’s distribution.
scipy.stats.dunnett() performs Dunnett’s test that does multiple comparisons of means against a control group.
from scipy.stats import dunnett
dunnett_res = dunnett(data_weed[data_weed['agent'] == '2']['yield_value'],
data_weed[data_weed['agent'] == '3']['yield_value'],
data_weed[data_weed['agent'] == '4']['yield_value'],
data_weed[data_weed['agent'] == '5']['yield_value'],
control=data_weed[data_weed['agent'] == '1']['yield_value'],
alternative='two-sided')
print(dunnett_res)Dunnett's test (95.0% Confidence Interval)
Comparison Statistic p-value Lower CI Upper CI
(Sample 0 - Control) 1.653 0.307 -0.068 0.304
(Sample 1 - Control) 2.143 0.129 -0.033 0.339
(Sample 2 - Control) 3.361 0.009 0.054 0.426
(Sample 3 - Control) 4.551 0.000 0.139 0.511The result shows (1, 4) and (1, 5) have different means. The figure below shows the CIs for
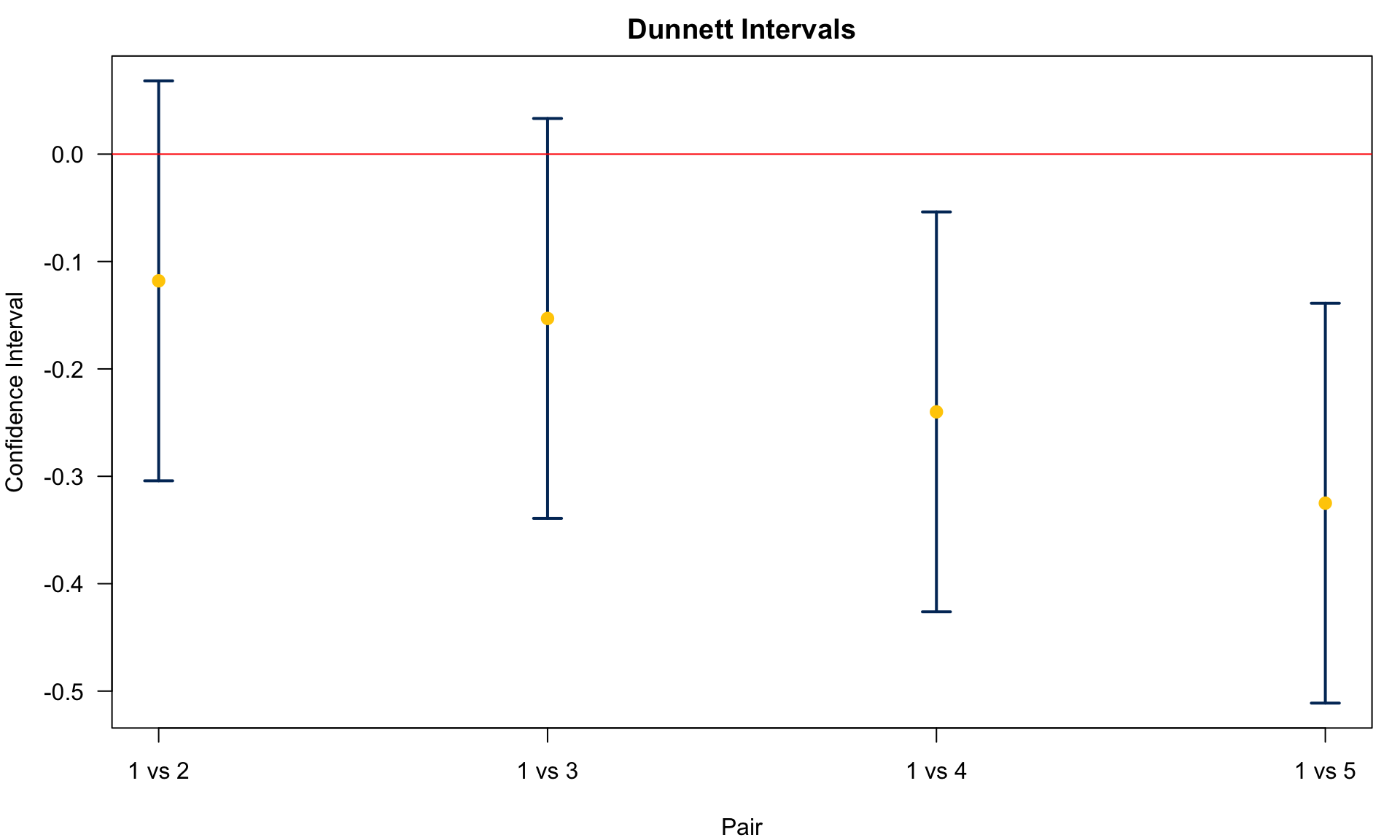
All comparisons illustrated are two-sided tests. One-sided tests can be applied too.
25.6 Comparison of Methods
To sum up, from the example, we had
Bonferroni:
Fisher’s LSD:
Tukey HSD:
Dunnett:
Fisher’s LSD does not control FWER, but all others do have FWER 0.05.
Bonferroni is the most conservative method and has the poorest discovery rate. You see its threshold value is the largest, making it the most difficult one to reject
Discovery rate for the Tukey’s is better than Bonferroni, but not as good as Dunnett’s. However, Dunnett’s used only to compare with the control.
If the objective is to compare only with a control, then Dunnett’s is more powerful among three. Otherwise, Tukey’s is more powerful than Bonferroni.
Although Bonferroni is not very powerful, it does have advantage that it can be used in any situation (whether it is one factor or multi-factor analyses) whenever there are multiple hypotheses.
All methods here are based on the condition that the data are random samples from normal distributions with equal variances. There are nonparametric multiple comparison procedures out there, such as Kruskal–Wallis.
Here we focus on balanced data. When there are large differences in the number of samples, care should be taken when selecting multiple comparison procedures.
-
There is another method called Scheffe’s method. Unlike methods specifically for pairwise comparisons such as Tukey HSD, Scheffe’s method can generally investigate all possible contrasts of the means. In other words, it considers not only th pairwise comparisons but any possible combinations of the means, for example
For pairwise comparisons, we reject