18 Inference About Variances
Why Inference for Population Variances?
We’ve learned how to do inference about one and two population means in the previous chapters. But we care about not only the center but also the dispersion or variability of some population distribution. In this chapter we are going to learn some inference methods for population variances. If the target population follows normal distribution
But why do we want to do inference about population variance? As population means, most of the time we do not know the true variance value, and in daily lives in some situations, we care about variation. For example, we would like to learn or control the variation in potency of some drug, say drug for lowering blood pressure. We hope the same amount or dose level of the drug provide the same effect on each individual. We don’t want some patients’ blood pressure is lowered a lot, but some other patient’s blood pressure is lowered just a little bit. In other words, we hope the drug effect is consistent or it has small variability. We want the new treatment can provide consistent potency and efficacy for all patients.
We also pay a lot of attention to the variance of stock prices because the higher the variance, the riskier the investment. We may want to monitor our financial portfolio, and the variability of our investment value so that we have long term financial stability.
Moreover, we may want to know if two population variances are equal, so that a correct or better method can be used. For example, in order to get higher statistical testing power, we use the two sample pooled
18.1 Inference for One Population Variance
Inference for Population Variances
Let’s start with inference for one population variance. For point estimation, the most intuitive and straightforward point estimator for
Note that the the denominator is
The inference methods for
The inference methods can work poorly if normality is violated, even if the sample is large.
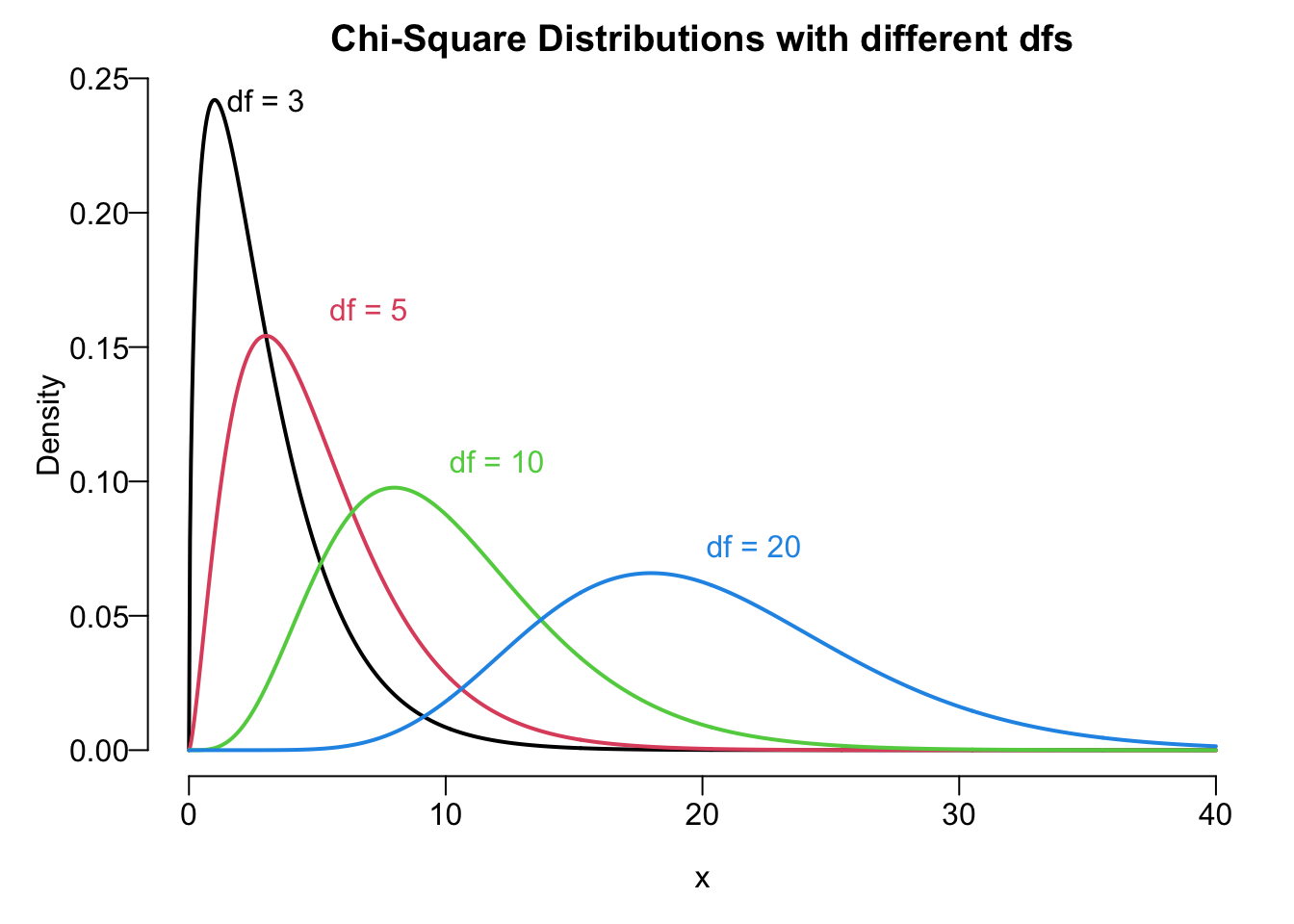
Chi-Square
Remember that when we do the inference for population means, we use either normal distribution or Student’s
As Student’s
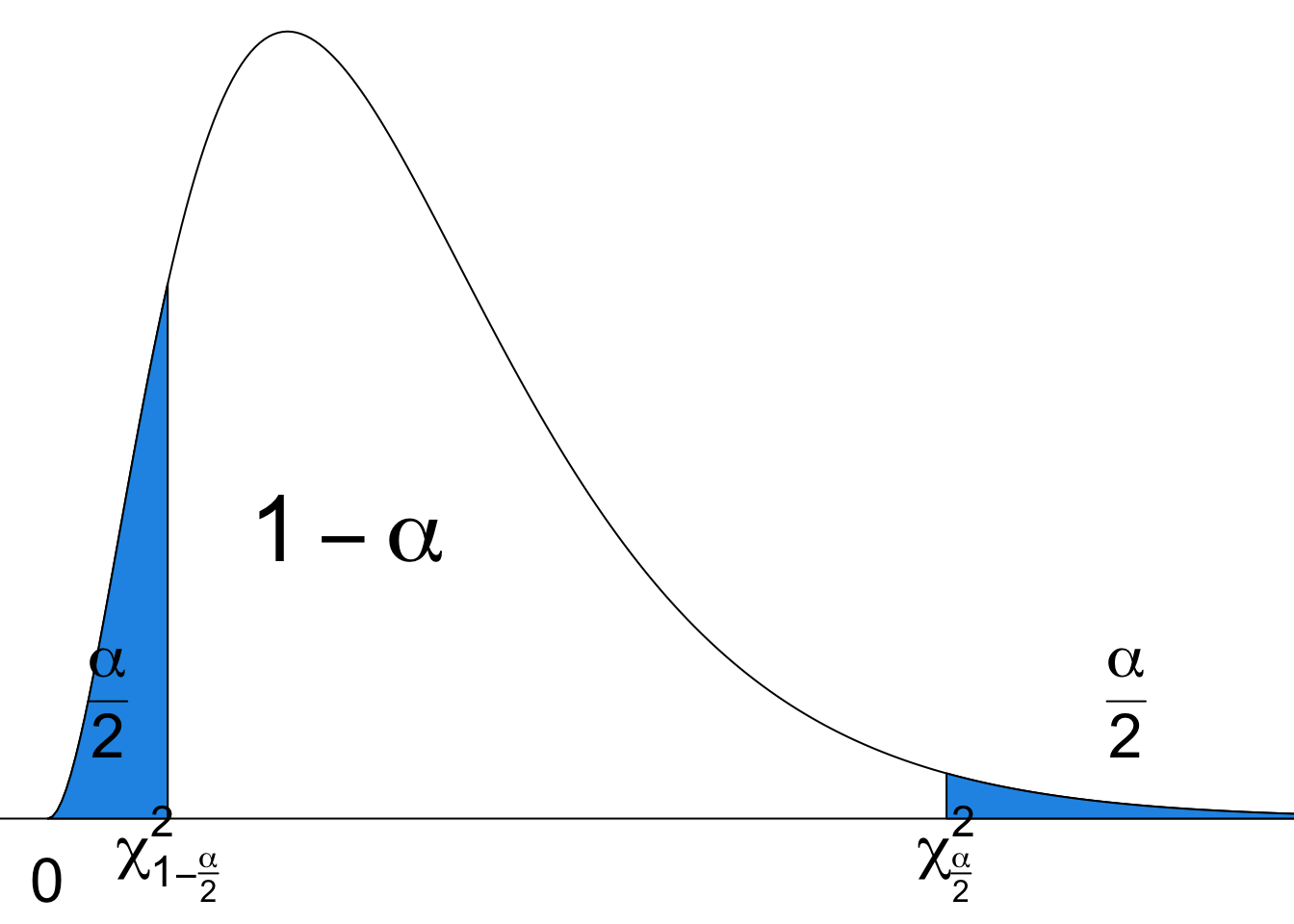
Upper Tail and Lower Tail of Chi-Square
Like standard normal and
Figure 18.3 illustrates the
Sampling Distribution
When a random sample of size
I would like to stress again that the inference method for
The sample statistic involves
The
How do we get this interval? We start with the sampling distribution of
-
-
-
-
Be careful that the interval for
Example: Supermodel Heights
Listed below are heights (cm) for the simple random sample of 16 female supermodels.
heights <- c(178, 177, 176, 174, 175, 178, 175, 178,
178, 177, 180, 176, 180, 178, 180, 176)import numpy as np
heights = np.array([178, 177, 176, 174, 175, 178, 175, 178,
178, 177, 180, 176, 180, 178, 180, 176])The supermodels’ heights are normally distributed. Please construct a

We just need to get what we need for constructing the interval from the sample data, sample size
-
Note that we want the interval for
We are 95% confident that the height of supermodels has standard deviation between 1.36 and 2.85.
We use qchisq() to get the df need to be specified. You should be able to understand the rest part of the code. Enjoy it.
## set values
n <- 16
s2 <- var(heights)
alpha <- 0.05
## two chi-square critical values
chi2_right <- qchisq(p = alpha/2, df = n - 1, lower.tail = FALSE)
chi2_left <- qchisq(p = alpha/2, df = n - 1, lower.tail = TRUE)
## two bounds of CI for sigma2
ci_sig2_lower <- (n - 1) * s2 / chi2_right
ci_sig2_upper <- (n - 1) * s2 / chi2_left
## two bounds of CI for sigma
(ci_sig_lower <- sqrt(ci_sig2_lower))[1] 1.362104(ci_sig_upper <- sqrt(ci_sig2_upper))[1] 2.853802import numpy as np
from scipy.stats import chi2We use chi2 family in scipy.stats to get the df need to be specified. You should be able to understand the rest part of the code. Enjoy it.
# Set values
n = 16
s2 = np.var(heights, ddof=1) # sample variance
alpha = 0.05
# Chi-square critical values
chi2_right = chi2.isf(alpha/2, df=n-1)
chi2_left = chi2.ppf(alpha/2, df=n-1)
# Confidence interval for variance (sigma^2)
ci_sig2_lower = (n - 1) * s2 / chi2_right
ci_sig2_upper = (n - 1) * s2 / chi2_left
# Confidence interval for standard deviation (sigma)
ci_sig_lower = np.sqrt(ci_sig2_lower)
ci_sig_lower1.3621044553698662ci_sig_upper = np.sqrt(ci_sig2_upper)
ci_sig_upper2.8538016067984957Testing
Back to the example. Use
Step 1
- We are comparing the
Step 2
Step 3
- The test statistic comes from the variable
Step 4-c
- This is a left-tailed test.
- The critical value is
Step 5-c
- Reject
- Since
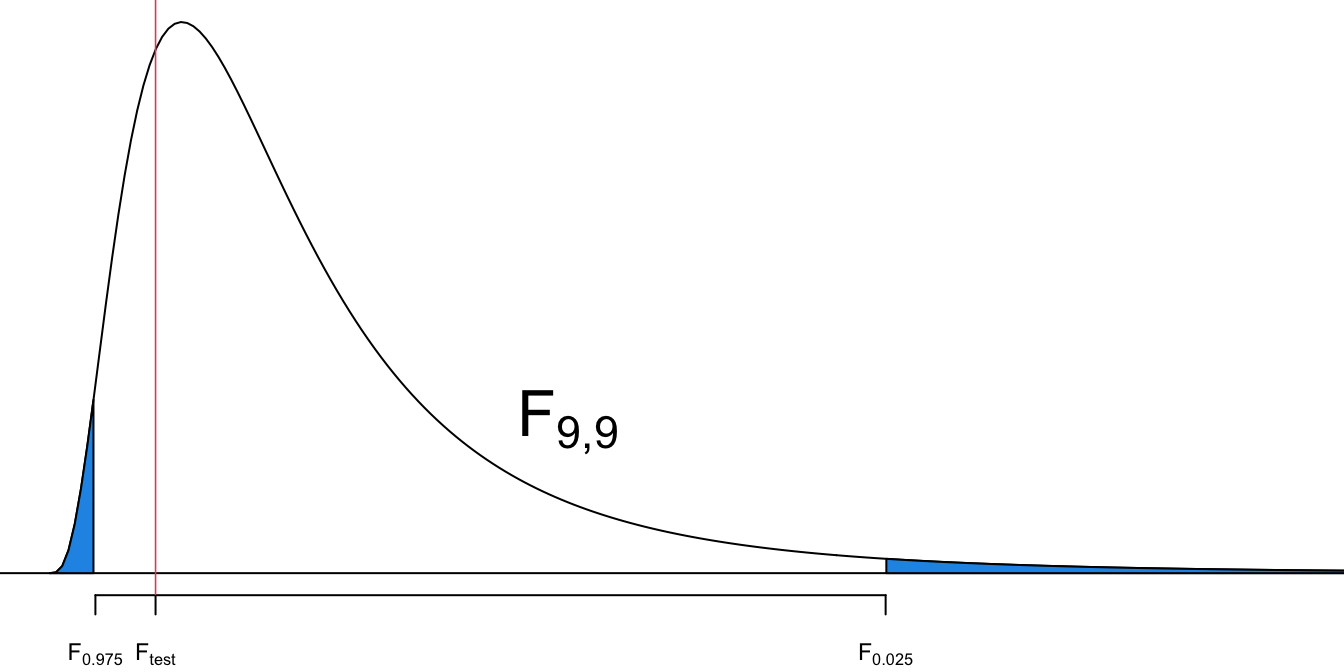
Step 6
- There is sufficient evidence to support the claim that supermodels have heights with a standard deviation that is less than the standard deviation for the population of all women.
We conclude that the heights of supermodels vary less than heights of women in the general women population.
Back to Pooled t-Test
In a two sample pooled t-test, we assume
-
-
We can use a QQ-plot (and normality tests, Anderson, Shapiro, etc.) to check the assumption of a normal distribution. We now learn how to check the assumption
18.2 Inference for Comparing Two Population Variances
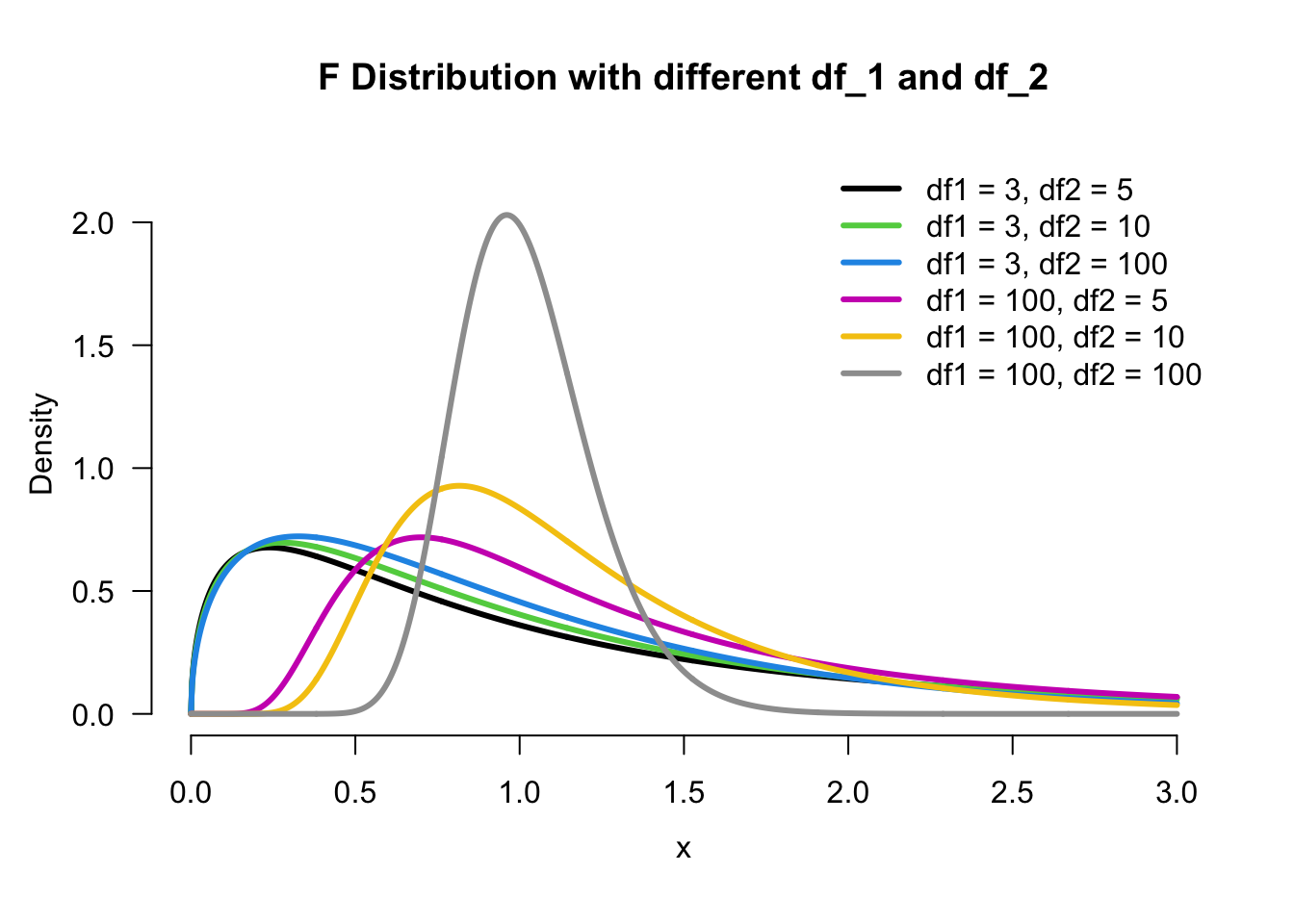
F Distribution
For comparing two population variances, we need another distribution called
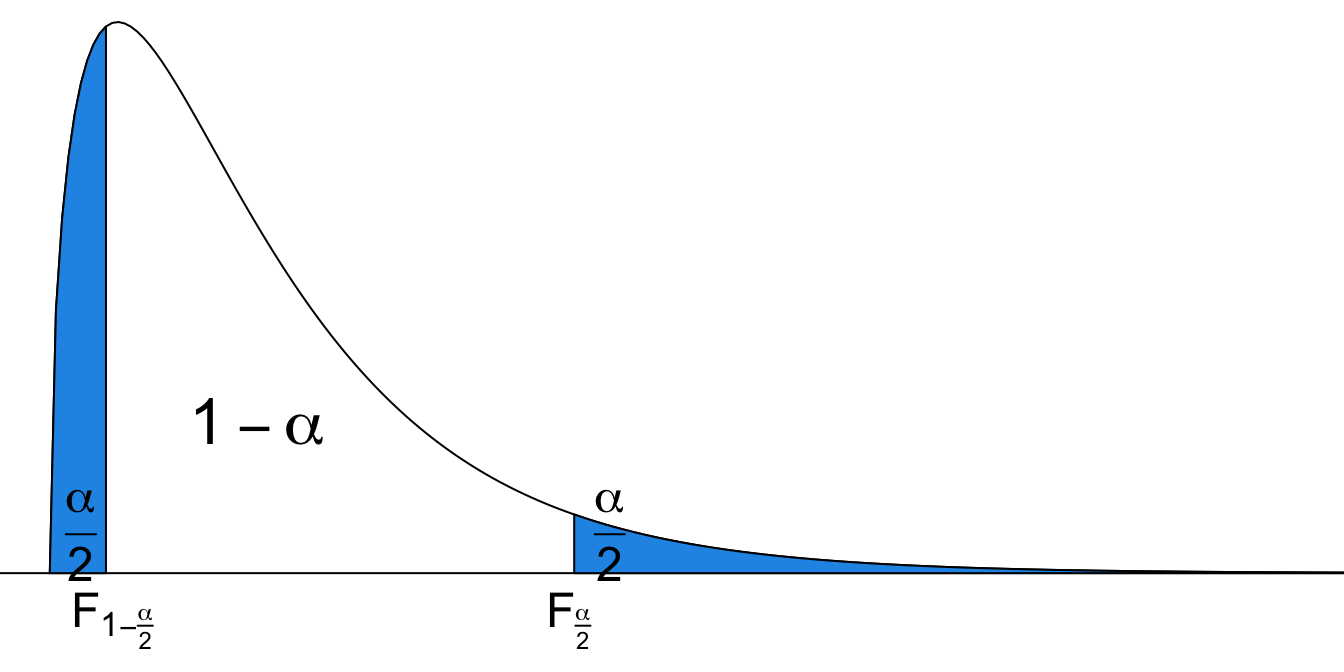
Upper and Lower Tail of F Distribution
We denote
When random samples of sizes
The order of degrees of freedom matters!
From the sampling distribution of the ratio, the
How do we get the interval? Not surprising. We start with the sampling distribution of
The confidence interval for
F test for comparing
Step 1
For comparing
- Right-tailed:
- Two-tailed:
Step 2
Step 3
- Under
Step 4-c
- Right-tailed:
- Two-tailed:
Step 5-c
- Right-tailed: reject
- Two-tailed: reject
Example: Weight Loss
This is our previous example.
A study was conducted to see the effectiveness of a weight loss program. Two groups (Control and Experimental) of 10 subjects were selected. The two populations are normally distributed and have the same standard deviation.

The data on weight loss was collected at the end of six months.
-
Control:
-
Experimental:
Check if
-
-
Step 1
Step 2
Step 3
- The test statistic is
Step 4-c
- This is a two-tailed test.
- The critical value is
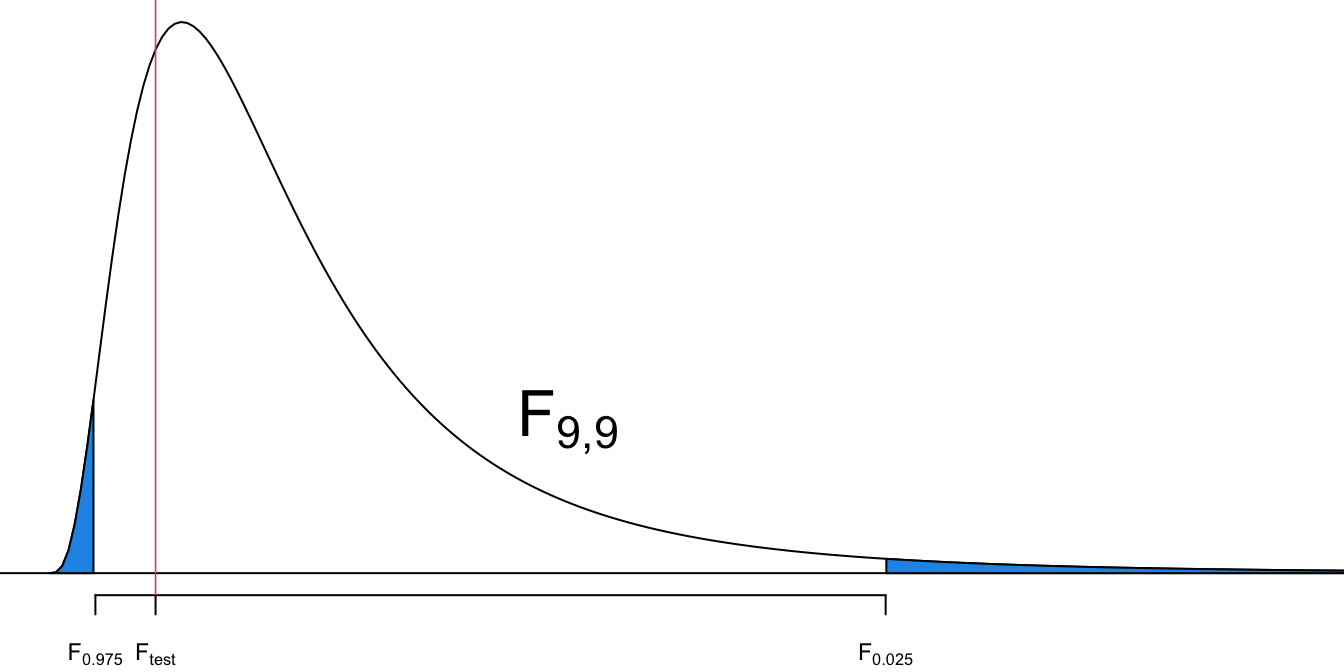
Step 5-c
- Is
- No.
Step 6
- The evidence is not sufficient to reject the claim that
95% CI for
- The 95% CI for
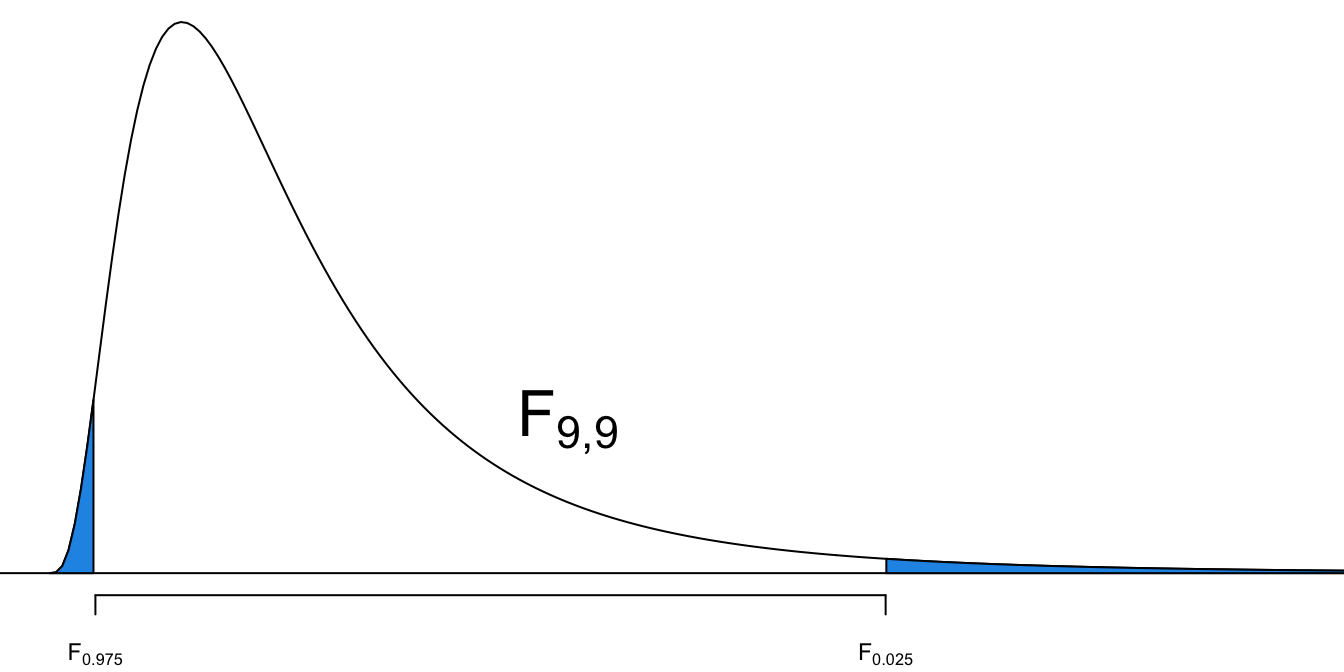
We are 95% confident that the ratio
We use qf() to find the
## set values
n1 <- 10; n2 <- 10
s1 <- 0.5; s2 <- 0.7
alpha <- 0.05
## 95% CI for sigma_1^2 / sigma_2^2
f_small <- qf(p = alpha / 2, df1 = n1 - 1, df2 = n2 - 1,lower.tail = TRUE)
f_big <- qf(p = alpha / 2, df1 = n1 - 1, df2 = n2 - 1, lower.tail = FALSE)
## lower bound
(s1 ^ 2 / s2 ^ 2) / f_big[1] 0.1267275## upper bound
(s1 ^ 2 / s2 ^ 2) / f_small[1] 2.054079## Testing sigma_1 = sigma_2
(test_stats <- s1 ^ 2 / s2 ^ 2)[1] 0.5102041We use f.ppf() and f.isf() in scipy.stats to find the
from scipy.stats import f
# Set values
n1 = 10; n2 = 10
s1 = 0.5; s2 = 0.7
alpha = 0.05
# F-distribution critical values
f_small = f.ppf(alpha/2, dfn=n1-1, dfd=n2-1)
f_big = f.isf(alpha/2, dfn=n1-1, dfd=n2-1)
# Confidence interval for the ratio of variances (sigma_1^2 / sigma_2^2)
# lower bound
(s1**2/s2**2)/f_big0.126727476884926# upper bound
(s1**2/s2**2)/f_small2.054078652185193# F-test statistic
test_stats = (s1**2)/(s2**2)
test_stats0.5102040816326532If we have the entire two samples data, we can use the R built-in function var.test(x, y, alternative = "two.sided") to perform a x and y are numeric vectors of data values. Argument alternative must be one of "two.sided" (default), "greater" or "less".
18.3 Comparing More Than Two Population Variances*
So far we use
where we have
Brown-Forsythe-Levene test actually represents two tests: Brown-Forsythe test and Levene’s test, where Brown-Forsythe test uses the sample medians in car::leveneTest() used in the example actually perform the Brown-Forsythe test by default with its argument center=median.
Using means has larger statistical power for symmetric, moderate-tailed, distributions. But when data are non-normal, either skewed or heavy tailed, using medians is recommended because of good robustness against many types of non-normal data and while retaining good statistical power.
The test statistic is
The decision rule is that we reject
There are other tests for testing the homogeneity of variances, but the BFL test performs better when data are from a non-normal distribution. It corrects for the skewness of distribution by using deviations from group medians. The BFL test is more robust. It is less likely to incorrectly declare that the assumption of equal variances has been violated.
Example: Automobile Additives (Example 7.8 in SMD)
Three different additives that are marketed for increasing the miles per gallon (mpg) for automobiles. The percentage increase in mpg was recorded for a 250-mile test drive for each additive for 10 randomly assigned cars. Is there a difference between the three additives with respect to their variability?
The data are saved in R data frame data_additive.
data_additive mpg_increase additive
1 4.2 1
2 2.9 1
3 0.2 1
4 25.7 1
5 6.3 1
6 7.2 1
7 2.3 1
8 9.9 1
9 5.3 1
10 6.5 1
11 0.2 2
12 11.3 2
13 0.3 2
14 17.1 2
15 51.0 2
16 10.1 2
17 0.3 2
18 0.6 2
19 7.9 2
20 7.2 2
21 7.2 3
22 6.4 3
23 9.9 3
24 3.5 3
25 10.6 3
26 10.8 3
27 10.6 3
28 8.4 3
29 6.0 3
30 11.9 3
The data are saved in Python pd.DataFrame data_additive.
mpg_increase additive
0 4.2 1
1 2.9 1
2 0.2 1
3 25.7 1
4 6.3 1
5 7.2 1
6 2.3 1
7 9.9 1
8 5.3 1
9 6.5 1
10 0.2 2
11 11.3 2
12 0.3 2
13 17.1 2
14 51.0 2
15 10.1 2
16 0.3 2
17 0.6 2
18 7.9 2
19 7.2 2
20 7.2 3
21 6.4 3
22 9.9 3
23 3.5 3
24 10.6 3
25 10.8 3
26 10.6 3
27 8.4 3
28 6.0 3
29 11.9 3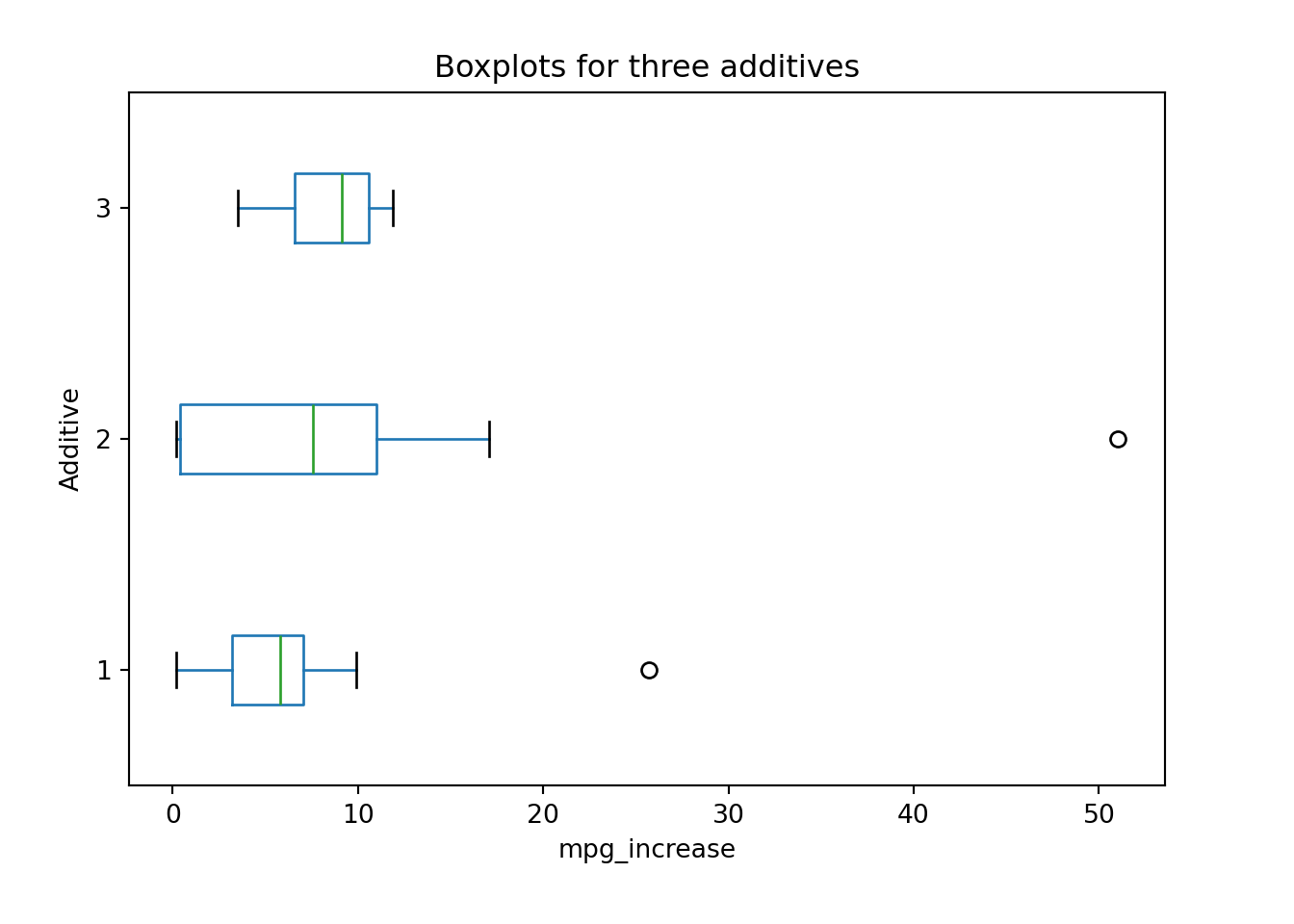
The boxplot suggests that Additive 2 has larger variation than the other two in terms of the percentage increase in mpg. We will use the BFL test to decide whether or not their variation difference is just a random sampling consequences or it is statistically significant.
The procedure of BLT test is shown step by step as follows.
Step 1:
Step 2:
Step 3: Test statistic
Step4-c: Critical value is
Step5-c: Since
Step6: There is insufficient evidence to support the claim that there is a difference in the population variances of the percentage increase in mpg for the three additives.
Once you understand the idea and the process of doing the test, in practice, we use R function leveneTest() in the car (Companion to Applied Regression) package to help us perform the test.
library(car) # Load the package car
leveneTest(y = data_additive$mpg_increase,
group = data_additive$additive)Levene's Test for Homogeneity of Variance (center = median)
Df F value Pr(>F)
group 2 1.8268 0.1803
27 In the function, we put the response variable data in y, and the factor defining groups in group. By default, it will use sample medians to perform the test. The p-value is greater than 0.05 and it leads to the same conclusion that there is insufficient evidence to say there is a difference in the population variances of the percentage increase in mpg for the three additives.
Once you understand the idea and the process of doing the test, in practice, we use Python function levene() in scipy.stats to help us perform the test.
from scipy.stats import levene
# Levene's test
levene(
data_additive[data_additive['additive'] == 1]['mpg_increase'],
data_additive[data_additive['additive'] == 2]['mpg_increase'],
data_additive[data_additive['additive'] == 3]['mpg_increase'])LeveneResult(statistic=1.82684271595955, pvalue=0.18025794484918528)In the function, each sample data can only be one-dimensional. By default, it will use sample medians to perform the test. The p-value is greater than 0.05 and it leads to the same conclusion that there is insufficient evidence to say there is a difference in the population variances of the percentage increase in mpg for the three additives.
18.4 Exercises
- The data about male and female pulse rates are summarized below.
- Construct a 95% CI for
- Construct a 95% CI for
- Does it appear that the population standard deviations for males and females are different? Why or why not?
- Construct a 95% CI for
| Male | Female | |
|---|---|---|
| 71 | 75 | |
| 9 | 12 | |
| 14 | 12 |